 Introduction to Machine Learning
Introduction to Machine Learning
- Resources
- Introduction
- Notation
- Classification
- Neural networks
- Regression
Preface
This website contains my lecture notes from a lecture by Ullrich Köthe from the academic year 2022/2023 (University of Heidelberg). If you find something incorrect/unclear, or would like to contribute, feel free to submit a pull request (or let me know via email).
The notes end at the decision tree/forest lecture (inclusive)!
Also, special thanks to Lucia Zhang, some of the notes are shamelessly stolen from her.
Resources
- TensorFlow playground – a really cool visualization of resigning and training a neural network for classification/regression and a number of standard parameters (activation functions, training rates, etc…)
Introduction
Setting:
- quantities that we would like to know but can’t easily measure
- quantities that we can measure and are related to
Idea:
- learn a function such that (we’re close to the real value)
Traditional approach:
- ask an expert to figure out , but for many approaches ML works better
Machine learning:
- choose a universal function family with parameters
- ex.
- find parameters that fit the data, so
Basic ML workflow:
- collect data in two datasets: training dataset (TS) to train and test dataset to validate
- extremely important, if you don’t do this you die instantly
- necessary for recognizing overfitting ( is polynomials and we fit points…)
- select function family
- prior knowledge about what the data looks like
- trial and error (there are tools to help you with this)
- find the best by fitting to the training set
- validate the quality of on the test set
- deploy model in practice
Probabilistic prediction:
- reality is often more complicated
- no unique true response , instead set of possible values (possibly infinite)
- in that case we learn a conditional probability (instead of a function):
- to learn this, we define a “universal probability family” with parameters and chose such that matches the data
- strict generalization: deterministic case is recovered by defining
- distribution has only one value
- often we derive deterministic results from probabilistic predictions (eg. mode/median/random sample of the distribution)
- generally, mode is the best, has some really nice properties
Problems:
- nature is not fully predictable
- quantum physics (randomness)
- deterministic chaos (pendulum)
- combinatorial explosion (aminoacids)
- data:
- finite
- noisy
- incomplete
- ambiguous
- model:
- function family is imperfect (too small)
- may not have converged
- nature has changed in the meantime
- goal:
- disagreement about what is relevant (desirable)
- handling uncertainty is central challenge of AI and ML
Notation
- features vector for each instance
- feature matrix (rows are instances, columns are features )
| (name) | (height) | (gender) | |
|---|---|---|---|
| Alice | m | f | |
| Bob | m | m | |
| Max | m | m |
We usually (when Programming) want a float matrix: drop names, discretize labels and use one-hot encoding (one-hot because there is only ones in the particular features).
| (height) | (f) | (m) | (o) | |
|---|---|---|---|---|
- response row vector for instance , response elements
- most of the time, (scalar response)
- exception is one-hot encoding of discrete labels
- tasks according to type of response
- is regression
- for number of categories classification
- labels are ordered (“tiny” < “small” < “medium” < “large”) – ordinal classification
- labels are unordered (“apples”, “oranges”) – categorial classification
- training set – supervised learning
- is the known response for instance
Types of learning approaches
- supervised learning – true responses are known in
- weakly supervised learning
- we have some information (know for some instances)
- we have worse information (there is a tumor but we don’t know where)
- unsupervised learning – only features are known in
- learning algorithm must find the structure in the data on its own (data mining or, for humans, “research”)
- only a few solutions that are guaranteed to work (this is unsurprisingly difficult to do)
- representation learning – compute new features that are better to predict
- clustering – group similar instances into “clusters” with a single representative
- self-supervised learning – define an auxiliary task where are easy to determine
Classification
-
- common special case , then or
- always supervised!
Deterministic classifier
- one hard prediction for each
Quality measured by collecting the “confusion matrix”
| true negative | false negative | |
| false positive | true positive |
- false positive/negative fraction – “how many out of all events are false”
- false positive/negative rate – “how many out of positive/negative events are false”
Probabilistic classifier
- returns a probabilistic vector (soft response) for every label posterior distribution
- more general than hard classification – can recover decision function by returning the most probable label by using :
Quality measured by “calibration”
- if , then label should be correct of the times
- if actual accuracy is higher, then the classifier is “under confident”
- otherwise it’s “over confident” (often happens)
Important: calculate confusion matrix or calibration from test set, not train set (bias)!
Threshold classifier
- single feature
- usually not enough to predict the response reasonably
- for some threshold
- three classical solutions for multiple features
- design a formula to combine features into single score (use BMI for obese/not obese)
- hard and expensive for most problems, not to mention inaccurate
- linear classification: compute score as a linear combination and learn coefficients
- less powerful than the first one (restricted to linear formulas)
- we can learn so it usually performs better
- nearest neighbour: classify test instance according to most similar training instance
- then
- expensive – to store the entire training set and scan it for every
- doesn’t scale well for more features (generalized binary search)
- hard to define the distance function to reflect true similarity
- “metric learning” is a hard task of its own…
- design a formula to combine features into single score (use BMI for obese/not obese)
Bayes rule of conditional probability
- joint distribution of features and labels
- can be decomposed by the chain rule to
- – first measure features, then determine response
- – first determine response, then get compatible features
- for classification, we want to use Bayes rule
- posterior – we want to update our judgement based on measuring the features
- prior – we know this (1% for a disease in the general population)
- likelihood – the likelihood of the disease given features (fever, cough)
- marginal – can be recovered by summing over possibilities:
Is hugely important for a number of reasons:
- fundamental equation for probabilistic machine learning
- allows clean uncertainty handling (among others)
- puts model accuracy into perspective (what is good or bad)
- defines two fundamental model types:
- discriminative model: learns (right side)
- answers the question “what class” directly
- (+) relatively easy – take the direct route, no detour
- (-) often hard to interpret how the model makes decisions (black box)
- generative model: learns and (left side)
- first learn how “the world” works, then use this to answer the question
- – understand the mechanism how observations (“phenotypes”) arise from hidden properties
- (-) pretty hard – need powerful models and a lot of data
- (+) easily interpretable – we can know why it was answered
- first learn how “the world” works, then use this to answer the question
- discriminative model: learns (right side)
- ex.: breast cancer screening:
- a test answers the following
- correct diagnosis:
- false positive:
- if the test is positive, should you panic?
- due to the very low probability of the disease, you’re likely fine
- a test answers the following
Some history behind ML:
- traditional science seeks generative models (we can create synthetic data that are indistinguishable from real data)
- physics understand the movement of an object, so a game can use this to appear real
- ~1930: ML researches realized that their models were too weak to do this field switched to discriminative models
- ~2012: neural networks solved many hard discriminative tasks the field is again interested in generative models (Midjourney, ChatGPT, etc.)
- subfield “explainable/interpretable ML”
How good can it be?
Definition (Bayes classifier): uses Bayes rule (LHS or RHS) with true probabilities
- we don’t know , we want to get as close as possible
Theorem: no learned classifier using can be better than the Bayes classifier.
- if our results are bad, we have to get better theory (more features)
How bad can it be?
- case 1: all classes are equally probable ()
- worst classifier: pure guessing – correct of the time
- case 2: unbalanced classes
- worst classifier: always return the majority label
Linear classification
- use a linear formula to reduce all features to scaled score
- apply threshold classifier to score ( for now)
for feature weights and intercept .
- learning means finding and
- we can also get rid of by one of the following:
- if all classes are balanced () then we can centralize features
- absorb into : define new feature (will then act as )
- done quite often ( is then called the “bias neuron”)
Perceptron
- Rosenblatt, 1958
- first neural network (1 neuron)
- idea: improve when model makes mistakes
- naive approach – 0/1 loss:
- can be used to evaluate but doesn’t tell us how to improve (and how)
- better idea – perceptron loss: weigh penalty by error magnitude (and use )
- note that these are only for one instance, for all the average error is
- naive approach – 0/1 loss:
Gradient descent looks as follows: for learning rate In our case, the derivative (for a single instance) is
Rosenblatt’s algorithm
Algorithm (Rosenblatt’s algorithm):
- initialzation of – random/uniform numbers
- for (or until convergence)
- update using the gradient descent formula above, with minor changes:
- additionally, use instead of (so it doesn’t change when learning set changes)
- sum over only incorrect guesses from the previous iteration
- update using the gradient descent formula above, with minor changes:
- only converges when the data is “linearly separable” (i.e. for loss )
- (+) first practical learning algorithm, established the gradient descent principle
- (-) only converges when the training set area linearly separable
- (-) tends to overfit, bad at generalization
Support Vector Machine (SVM)
Improved algorithm (popular around 1995): (Linear) Support Vector Machine (SVM)
- maintain a safety region around data where solution should not be
- closest points – support vectors
- learns , which is LHS of Bayes – is discriminative
Reminder: distance of a point and plane is
- notice that scaling doesn’t change the distance – pairs define the same plane – we can therefore define an equivalence class for representatives (can be chosen arbitrarily)
Radius of the safety region (margin) is the smallest distance distance of a point to the decision plane (also called the “Hausdorff distance between H and TS”):
Let be the closest point. Then
- we don’t use minus because we want distance, not penalty (like perceptron)
- we again use the trick with multiplying by the label and brought the norm to the other side
Now we chose a representative such that the equation above is . The decision plane is then correct when (specifically for ). To make it as general as possible, we want one that maximizes the distance
This yields an optimization problem: we want to
This is inconvenient – change to a more convenient equivalent optimization problem
Now we define slack variables that measure how much it was violated and we also want to minimize those, which we’ll do by using Lagrange multipliers:
- adjusting makes compromise between being right vs. robust (hyperparameter tuning):
- approach: 3 datasets – training, validation, testing
- pick a set of possible hyperparameters ()
- train with each on the training set
- measure accuracy on the validation set
- keep the best on validation set, test again on the test set
- validation is present because test set would otherwise be training too…
- approach (2 datasets): cross-validation
- split training set into pieces (“folds”, typically around or )
- has to be randomly ordered, otherwise it won’t work!
- use each fold as validation set in turn & train on the remaining folds
- special variation: -fold (train on everything besides one instance)
- expensive in practice, great in theory
- split training set into pieces (“folds”, typically around or )
- approach: 3 datasets – training, validation, testing
- comparing with perceptron:
- we introduced (no normalization)
- we now have a regularization term (no overfitting!)
- we have : if you’re barely right, you still pay small penalties!
We again optimize by derivative + gradient descent:
The iteration step for looks as follows:
- note that we can’t get stuck in a minimum, since the objective function is convex
Linear Discriminant Analysis (LDA)
- idea: assume that features for each class form a cluster
- assume they’re elliptic and model each one as a Gaussian distribution
- is RHS of Bayes (generative) – calculate the posterior from Bayes formula
- Linear – clusters have the same shape
- there is also a Quadratic, which permits different shapes but isn’t linear
To get any ellipse, we start with a unit circle and stretch (), rotate () and shift () it:
Solving for (and using the fact that is orthogonal), we get
Gaussian distribution can be defined as
- for mean, variance
- standard normal distribution is for and
For higher dimensions (with our circle ), we get the generalized Gaussian
- is the precision matrix
- since , we see a decomposition to eigenvalues (correspond to square of radii of the ellipse) and eigenvectors (the corresponding radii vectors)
Let be features of class . Then
- learning: find and
To derive the learning method, we’ll use two things:
- maximum likelihood principle: choose and such that TS will be a typical outcome of the resulting model (i.e. best model maximizes the likelihood of TS)
- i.i.d. assumption: training instances drawn independently from the same distribution
- independently – joint distribution is the product
- identically distributed – all instances come from the same distribution
For the probability, we get
Using the maximum likelihood principle, the problem becomes
It’s mathematically simpler to minimize negative logarithm of (applying monotonic function to an optimization problem doesn’t change and ). We get
This is our loss. We now do derivative and set it to , since that will find the optimum. First, we’ll derivate by , which gets rid of the first part of loss completely and we get
In other words, the mean is the average (shocking, I know).
For , this will be a little more complicated. We’ll do the partial derivation by instead, which gives the following:
Again, in other words, the variance is the average over the quared vectors offset by the mean, which too makes sense.
Now we have clases but with same covariance (by assumption of LDA) and we can:
- determine two means () as
- to calculate covariance (which is the same for both classes):
- use Bayes RHS and our calculations to calculate the LHS:
The derivation of (3) looks as follows: assuming for simpler computations and being analogous:
Now we substitute the Gaussian function:
Plugging the result into the previous equation, we obtain the sigmoid function :
Two of its properties will be helpful for us:
Doing an analogous derivation for , we get
Alternatives to training beta and b:
- fit mean and covariance of clusters
- least-squares regression: (same solution as )
- Fisher’s idea: define 1D scores and chose such that a threshold on has minimum error
- define projection of the means
- intuition: and should be as far away as possible
- doesn’t quite work, because
- solution: scale by the variance : , then we get
- again gives the same solution as and
- Logistic regression (LR): same posterior as LDA, but learn LHS of Bayes rule
- gives different solution to LDA
Logistic regression (LR)
We again have i.i.d. assumptions – all labels are drawn independently form the same posterior:
- as a reminder, this was swapped for LDA (we had )
- use maximum likelihood: choose such that posterior of TS is maximized:
For LR, , which allows us to rewrite (with the results from LDA) like so:
- no analytic solution but is convex (local extreme = global extreme, solve via GD)
Here we have , which is called the softplus function:
Simplifying for and using the following properties:
We get the derivatives :
obtaining
The four cases that can occur are as follows:
| Real value \ Classifier result | ||
|---|---|---|
| no correction | error : pulls | |
| error : pulls | no correction |
Summary
- with , all methods have same decision rule but the methods differ by how they define & find optimal
- common objective function
Perceptron:
SVM:
LDA:
LR:
- in practice, similar solutions when data is linearly separable
- different tradeoffs otherwise check validation set
Multi-class classification
- solution: reduce to a set of -class problems
- one-against-rest classification
- for , define score
- train by treating as “class + 1” and as “class - 1” (rest)
- make them comparable by normalization: (same for )
- classify according to biggest score, or “don’t know” if all scores are bad:
- all-pairs: train a linear model for all ()
- if then one vote for class , if then one vote for
- is the label with most votes (or “unknown” for ties)
- define posterior as “softmax-function” scores as in (1):
- standard for neural network classification
- easily provable:
- new: train all jointly (at the same time)
Non-linear classification
- in practice, use LR and hope for the best
- exception: very scarce data hard to justify non-linear fit
- measure more features and increase dimension
- higher dimensional spaces tend to be more linearly separable
- for , it’s always separable (but maybe not useful)
- for , use sparse learning methods
- use non-linear classifier
- QDA, which is a non-linear version of LDA
- Kernel-SVM (non-linear SVM, less popular but cool math)
- Decision trees/forests (recursively subdivide , we’ll discuss them later)
- non-linearly transform features
- XOR function: ,
- four points classification:
- BMI (non-linear formula, linear classification)
- problem: hand-crafting is difficult
- solution: learn (multi-layer neural networks)
Neural networks
- definition: inputs eg. ( or output of other neurons)
- computation (activation): (linear function plugged into non-linear one)
- pre-activation:
- popular activation functions:
- identity: used as output of regression networks
- classic choices: or , almost the same when scaled / offset
- modern choices:
- – not differentiable at but nice in practice
- leaky
- exponential linear unit
- swish function
- usually, is not used but turned into a “bias neuron” for each layer
A neural network is a collection of neurons in parallel layers
- fully-connected: all neurons of previous layer connect to those of the next
- for layers, we have (bias), hidden and
- sample computation would then be
- (1) is the input, (2-5) are hidden layers, (6) is the output
Previously, NN were believed to not be a good idea, but
- ~2005: GPUs were found out to compute them well
- big data became available (to train the big network)
- ~2012: starts outperforming everything else
Activation functions:
- (not for input): chosen by the designer
- nowadays we usually use one of the modern choices described above
- : determined by application
- regression (): identity
- classification ():
Important theorems:
- neural networks with ( hidden layer) are universal approximators (can approximate any function to any accuracy), if there are enough neurons in the hidden layer
- purely existential proof, but nice to know
- up till 2005, this was used, since they were enough
- bad idea – deeper networks are easier to train (many good local optima)
- finding the optimal network is NP-hard we need approximation algorithms
Training by backpropagation (chain rule)
- idea: train by gradient descent need
- update is the same as previous GD:
- to compute the derivative, we’ll compute the chain rule from last layer backwards
- (application-dependent loss)
- regression:
- classification:
- back-propagate through output activation
- here we define since we’ll use it a lot
- for regression, we had identity activation function so
- for classification, this is a bit more complicated but ends up being
- for every , let ; recursion starts with (previous step)
- use chain rule (and simplify/compute using previous layers):
- another chain rule, finally with what we want to calculate:
Training algorithm:
- init randomly (explained later)
- for
- forward pass – for in batch
- for , compute + store along the way
- backward pass: , for in batch
- compute
- for , compute
- update:
- forward pass – for in batch
Convolutional Neural Networks (CNNs)
- previously, we had fully connected networks
- work well for ~1000-dimensional data (can handle MNIST)
- CNNs are typically used for images, neurons are sparsely connected
- every CNN can be implemented as a (much larger) fully connected network
- neurons are only connected based on its top (see below)
- idea: objects in images don’t change when at different location
- classification should be translation invariant
- the learned features should be translation equivariant
- when dog moves, features move in the same way
- these are two of the defining ideas of a convolution
Definition (filter): consumes an image and outputs another “filtered” image
For our filter, we want translation equivariance and linearity
Theorem: any such filter can be written as a convolution integral, i.e.
Substituting , we see that the operation is commutative:
Since we don’t have infinite images, , the integral becomes a sum:
Trick: if we want to find of a black-box filter, take (Dirac delta function), then
Usually, has a finite support, i.e. if , then we get
- has non-zero elements so it’s called the -tap filter
- 3-tap filter (in 2D )
- 5-tap filter (in 2D )
Why is this useful? – “matched filter principle”
- is big if around is similar (“matched”) to
- the kernel of the filter encodes the pattern of interest (matches similar features)
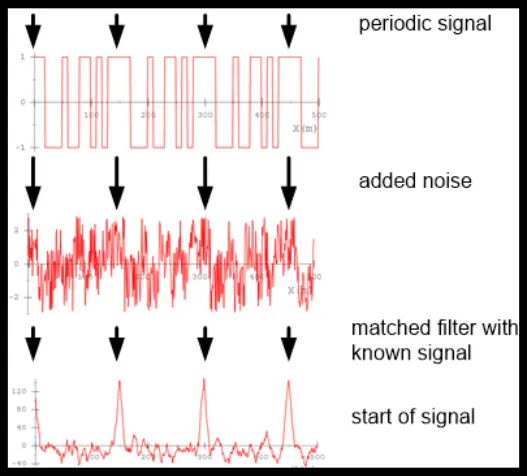
If we repeat this in multiple layers and learn the s, we can do very powerful things.
Fun fact: in practice, CNNs do not implement convolution but instead correlation for maths reasons (the maths requires the mirrored convolution, which is correlation).
To build a CNN (3-top):
- each neuron only connects to 3 neurons of previous layer
- image boundaries have to be resolved:
- cut off the offending neurons
- zero padding
- reflect boundaries
- image boundaries have to be resolved:
- weights are shared in each layer based on if the connection is left/middle/right
- high response for the given pixel is a match
Example: LeNet architecture (Yann LeCun, 1998, 7 layers)
- first CNN to get MNIST accuracy > 99%
- ~60000 images, 10 categories
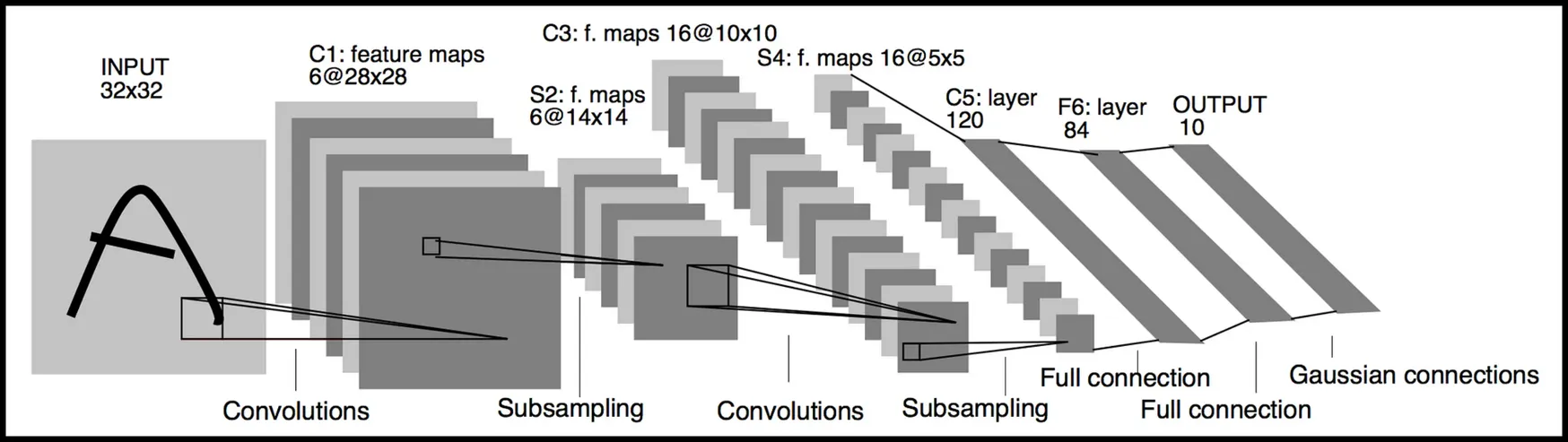
Comparing a CNN to a fully-connected (FC) network, the weight matrices look like this:
Here we handle the boundary with zeroes, but it can be done differently:
- for reflected, the leftmost would be (along with the rightmost )
- for periodic, the rightmost top cell would not be but instead (+ some others)
Here we’ve seen one of the major operations, convolution, the other being pooling:
- reduces the image resolution, done by merging several input pixels into one output pixel
- typically reduces
- not sliding anymore, move by the window size instead (running window)
- usually either average pooling or max pooling
CNNs usually alternate between convolution, non-linear layers (ReLU), pooling, dropout layers, batch normalization layers and (towards the end of the network), fully-connected layers and softmax layers.
Receptive field of a neuron
- set of input pixels (from original image) that influence the value of a given neuron
- i.e. maximum size of detectable patterns
- RF of:
- one convolutional layer is the size of the kernel
- sequence of convolutional layers is additive (wrt. kernel radius)
- pooling is multiplicative (wrt. pooling size)
Some history
- ImageNet (2008) ~14 mil. images, more than 1000 classes (224x224x3)
- 1st public challenge (2010)
- predict 5 guesses for the object class
- correct if true label is within top 5 (“top-5 acc.”)
- winner: non-linear SVM with hand-crafted features (28% top-5 error)
- 2nd public challenge (2012)
- winner: CNN AlexNet (15.3% top-5 error)
- 3rd public challenge (2014)
- winner: GoogLeNet – inception architecture (6.7% top-5 error)
- runner-up: VGG – upscaling of LeNet (7.3% top-5 error)
- clean and popular (compared to winner, which was quite hacky)
- used as a comparison metric for things like Midjourney/DALL-E (FID score)
- making VGG bigger did not work (vanishing gradient problem) – magnitude of gradient at the prompt decreases with the number of layers and the network doesn’t converge
- 2015: Residual Network (ResNets)
- instead of learning a function per layer, learn weights wrt. the input layer
- also skip connections as identity mappings, preventing vanishing gradients
- reaches superhuman performance (humans 4/5%, ResNet 3.6%)
- for top-1, we they had 25% error (nowadays ~10%)
- nowadays, use two tricks:
- train on much larger datasets (up to 3B images)
- 1st public challenge (2010)
Residual Networks (ResNet)
- base idea: use skip connections to avoid the vanishing grandient problem
- stages between pooling layers (image resolution unchanged within the stage)
- block – sequence of layers + one skip connection (bridges the block):
- batch normalization: adjust mean and variance of pre-activations
- bad when pre-activations have an arbitrary mean and variance (data should be within the intersting region of the activation function)
- () calculated for each batch
- batch normalization: adjust mean and variance of pre-activations
- layers – do the actual work
Training
- rule of thumb: more data is better (nowadays images)
Data augmentation
- apply transformations to the data that keeps the classification unchanged
- artificially increases the dataset
- popular augmentations:
- geometric distortions:
- mirroring, 90deg rotations
- shearing % small angle rotations
- crop regions & resize to original size
- image quality distortions:
- add noise – Gaussian / salt & pepper (random 0/1 pixels)
- mash out regions (set to gray)
- color distortions:
- transform to gray
- transform color space (eg. LAB)
- other distortions:
- edge detection (human should still recognize it)
- geometric distortions:
- hyperparameters:
- amount of distortion
- number of simultaneous distortions ( works good)
Self-supervised training
- use augmentation to avoid manual labelling
- strategy (1): use augmentation that can be labeled automatically
- rotate by predict
- to solve this, network learns features that are useful for other tasks – cut out the angle detection and use the reset of the network for feature detection
- strategy (2): contrastive learning
- present augmented pairs; if it originated from the same image, features should be similar, else they should be different (ie. ignore data augmentations)
- SIMCLR network – famous for doing this
- strategy (3): semi-supervised learning
- subset of the training set is labeled and rest is unlabeled
- student-teacher approach:
- train the teacher from the labeled and then use the teacher to label the unlabeled (preferably soft labels since it can make mistakes and might not be sure)
- train the student with the original and the pseudolabels
- experimentally, often the student is better / has a smaller network
U-Net
- tasks in image analysis:
- classification (what we were talking about), 1 label per image
- instance classification & segmentation, 1 label + bounding box per instance
- semantic segmentation, 1 label per pixel (one-hot vector per pixel)
- less difficult than (2), since objects can be occluded by others
- U-Net solves (3)
- has the name because it looks like the letter U
- idea: coarsening to detect the semantics + refinement for pixel-precision
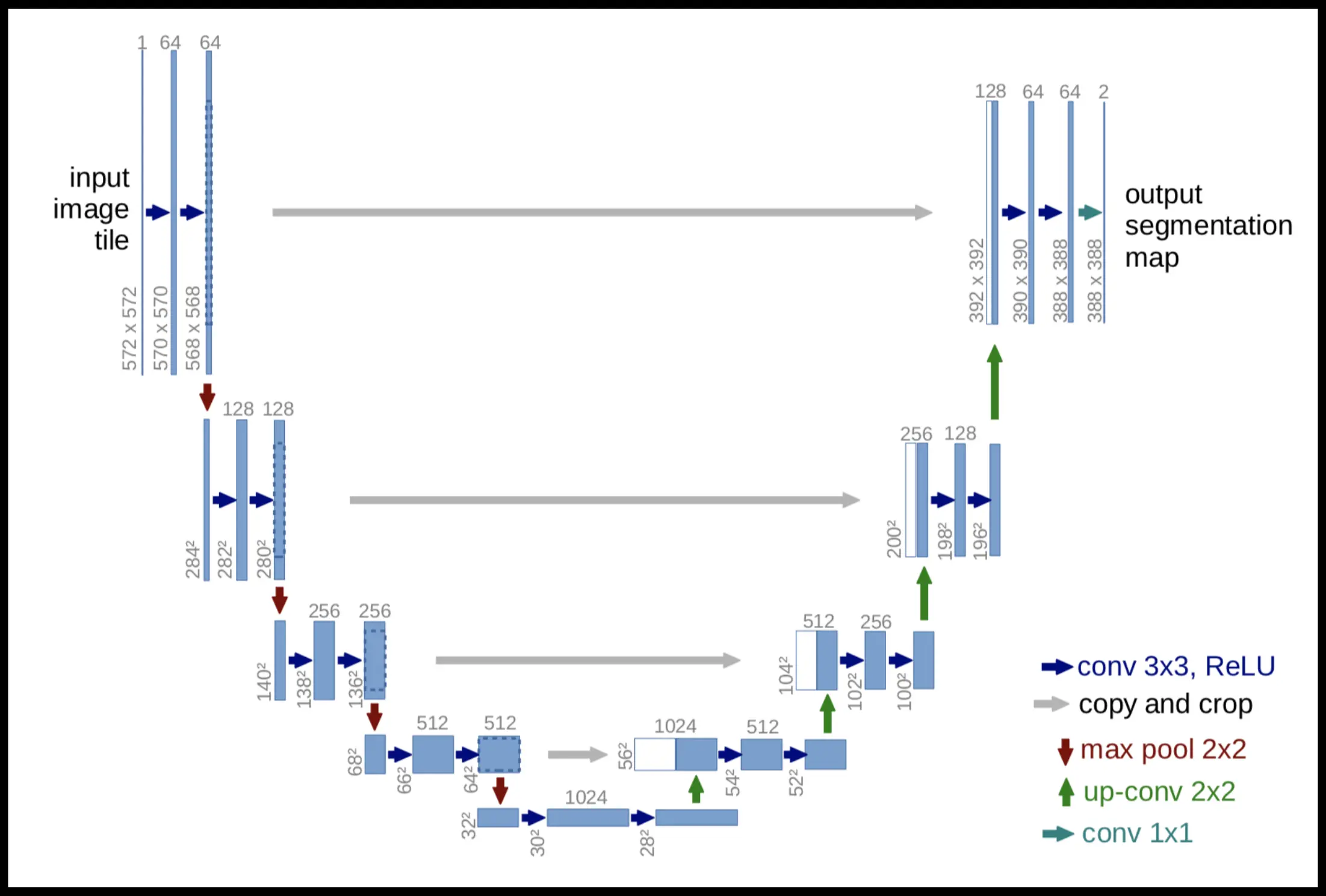
- variation: nnU-Net (2021)
- designed here in Heidelberg!
- previously, people designed new net for every task
- observation: have rules to design the network given the task (nn = no new)
- won about 30 medical benchmark competitions
- U-net is a specific example of an auto-encoder:
- takes in a -dimensional data
- then uses an encoder to convert to -dimensional codes
- lastly uses a decoder to convert back to the ~original data
- since , this is lossy, but works really well
- we want to minimize
Regression
- features , response real-valued scalar (or vector)
- compared to classification where is discrete
- predict deterministically () or stochastically ()
- functional representation lemma (aka “reparametrization trick” / “noise outsourcing”)
- every conditional probability can be expressed by a deterministic function & independent noise: with drawn from a distribution (usually Gaussian) independent from ; eg.
- much more convenient since the distribution is fixed (not dependent on )
Linear regression
- easy case, just like linear regression
- assume that data is generated by some true (but unknown) generative process
- in this case linear model with additive Gaussian noise, for simplicity assume (i.e. variance is fixed but unknown)
- given , find and
- important: assume that only is noisy, not (other variant also exists)
-
derive loss by maximum likelyhood principle and i.i.d. assumption:
- squared loss ordinary least squares (OLS) regression
- can be solved analytically:
Written another way, we get meaning that the regression always goes through the center of mass of TS, which allows us to first center the data (), making the center of mass the origin and getting rid of (same as classification).
Now we want to generalize to feature vectors – (row vector) rewrite the objective in matrix form (still assume centered data):
where , so we get
which are called the normal equations of the OLS problem, since is the normal of the regression plane and we can solve it as
Pseudoinverse is a generalization of the inverse for rectangular matrices:
- not a great way to calculate this – expensive and also might be too degenerate
- if at least one feature (column of ) can be expressed as a linear combination of other features, it will be entirely degenerate and OLS will have no solution
- contition number measures this:
- if is nice (almost orthogonal features)
- if has almost redundant features, becomes inaccurate
- if has redundant features, doesn’t exist
- infinity arrises from the norm of (division by )
We can instead solve OLS by SVD:
- every matrix can be decomposed into for
- orthonormal matrix
- diagonal matrix
- quasi-orthonormal matrix
- using this we get
- if features are redundant, eigenvalues are
- when we’re doing , this explores, set
- this gives us the minimum norm solution of the degenrate OLS problem
- condition number for SVD is
- advantage: numerically very stable, even when is almost degenerate
- disadvantage: very complicated algorithm (500 LOC of Fortran); nowadays this is ok
Third solution: QR decomposition
- much easier than SVD, reasonably robust for bad data
- uses a different decomposition for
- orthonormal
- upper triangular
- using this, we get which can be solved by a simple algorithm – is upper triangular so we can use substitution from the last row of upward
What do we do if and are very big?
- in theory, we’re screwed (SVD complexity )
- in practice, big matrices usually have special structure which we can exploit
- sparse – alg. only deals with the non-zero elements
- is a convolution – even more strict
- we don’t access directly, but via subroutines
- vector-matrix product
- matrix-vector product
- all the tricks to exploit the special strucure of hidden here:
LSQR
- trick: only use the subroutines to exploit the structure
- trick: rearrange the computation such that only 2 rows or columns of the result matrices are needed simultaneously (in memory)
- LSQR decomposition is where
- is orthonormal
- is upper bi-diagonal
- diagonal + one more diagonal above are non-zero, the rest is zero
- is orthonormal
- implemented as
scipy.sparse.linalg.lsqrin Python (or new and improvedlsmr)- arguments are sparse matrices
The lecture here has an interlude into computer tomography, which can be solved using least-squares. Most of it is explained in homework 4 so I’m not going to repeat it here.
Least Squares v2.0
What do we do when noise variance is not constant?
- i.e. we change the generative model into
- data still independent but no longer identically distributed
- we use since we want to learn both and
Here we distinguish multiple cases:
- OLS (all s are the same) – we can simplify further and get stuff from lectures above
- weighted LS ( are known) – then
- the s act as weights for the residuals
- we can rewrite in matrix notation (s as a diagonal) and get
- this can be solved by setting the derivative to zero and we get a weighted pseudo-inverse
- iteratively reweighed LS ( are not constant and unknown) – learn the s jointly with )
- is unsupervised, is supervised – gives rise to interesting algorithms
- the problem can be formulated as usually called David-Sebastian score or hetero-scedastic loss
- if is big increase to make the loss smaller, but we pay the penalty of for big s (optimal solution selects for the best trade-off)
Alternating optimization
- we want to solve IRLS by alternating learning and
- two groups of parameters (here )
- main idea: optimize while keeping fixed (and vice versa)
- define initial guess for
- for (or until convergence)
- optimize , keeping fixed
- optimize , keeping fixed
- benefit: individual optimization problems are much easier (can have an analytic solution)
- drawback: usually doesn’t converge to the global optimum
To solve IRLS using this method, we do the following
- define initial guess as (
- for (or until convergence)
- obtain via weighted least squares (since s are known and fixed)
- since is fixed, we get which we can solve by setting the derivative to and obtain i.e. standard derivations are equal to the magnitude of the error
How many iterations are required?
- in theory (with infinite accuracy) two iterations are sufficient
- in practice few iterations are sufficient (kinda cool, no?)
Regularized regression
- even if we don’t have a unique solution, we can select the “best” one
- we already saw penalties in SVM and in IRLS
- we need regularization in two cases
- more features than observations ()
- condition of matrix is bad ()o
- features are almost redundant, tends to overfit
- assume feature and are identical in TS ()
- if is a solution, so it with and
- this also works for very large , which is a problem since the features might be different in test instances (i.e. overfitting)
- in practice, usually more features and only near cancelation
Ridge regression
Uses a standard regularization idea to penalize with large magnitude (or squared magnitude):
- if constraint doesn’t activate after we solve, we’re chilling
- otherwise add the constraint as a Lagrange multiplier (same as SVM)
Setting the derivative to zero, we obtain the regularized pseudo-inverse
- data should ideally be standardized (zero mean, unit variance) so we shrink equally
Alternative solution via augmented feature matrix – reduces to OLS by modifying and to be
- by expanding OLS, we can see this is equivalent to original loss
Another alternative solution is via SVD.
Bias-variance trade-off
When using regularization, we’d like to know two things:
- what’s the price that we pay for regularization?
- how to choose to minimize disadvantages
Both solved via bias-variance trade-off: we have teams working on the same problem, each with their own :
- how much will the results differ between teams?
- what happens when ?
Definitions:
- : weights of the true generative process
- : results of team
- : average result of all teams
- :
- : systematic error after combining team results
Now we’ll measure the quality of outcomes by
Useful, because we can see that (regularization) has opposite effects on bias and variance – “bias-variance trade-off”
- high variance (teams disagree) but zero bias
- all teams same solution, but it’s
- agreement between teams increases but so does bias (trade-off)
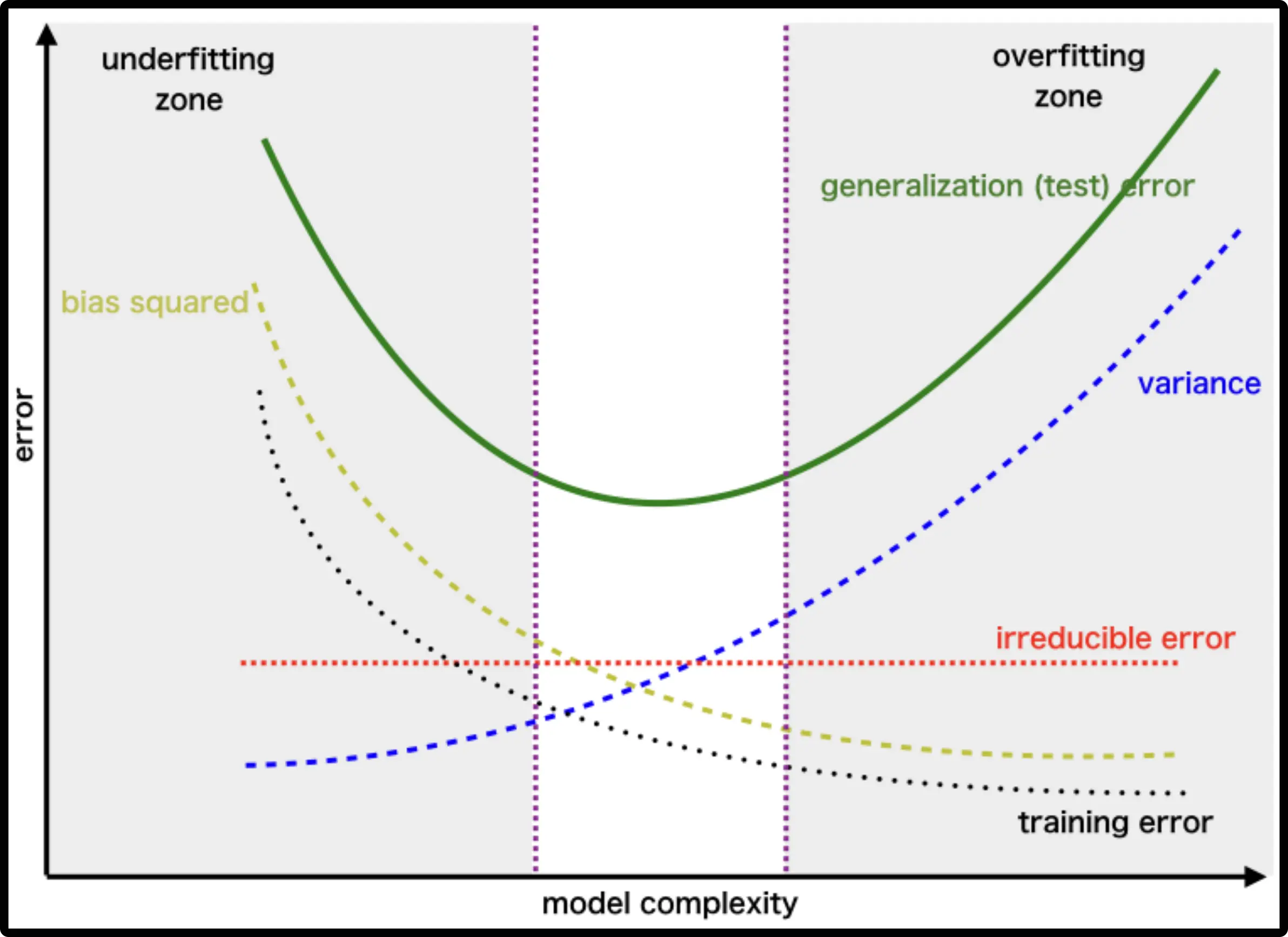
- best trade-off: all error sources have roughly same magnitude
- check via cross-validation (or validation set)
- bad: only address a single error source (diminishing returns)
Other regularization terms
- Ridge regression: regularization term
- Feature selection:
- count how many features are active
- advantage: reduce the number of measurements
- disadvantage: finding the optimal feature set is NP-hard (but in practice a simple greedy algorithm like OMP is often good enough)
- LASSO regression:
- stands for Last Absolute Shrinkage Selection Operator
- also performs feature selection when is small enough
- advantage:
- still a convex optimization problem unique solution
- solution often sparse (many ) feature selection
- disadvantage:
- bias (bias-variance trade-off)
- active set may not be as sparse as in optimization
- many algorithms (in the field of linear optimization)
- gradient descent
- LARS (Least Angle RegreSsion) algorithm – almost the same as OMP, but at the beginning of each iteration, checks if the matrix is redundant (and removes weakest factors until it isn’t); doesn’t happen too often though
Orthogonal Matching Pursuit (OMP)
- approximately (local optimum) solves feature selection, i.e.
- initialization: set of active features
- for (max. number of features)
- compute residual of current guess
- find best inactive feature
- if the direction of the new feature is similar to the residual, it improves the solution by a lot
- if it’s orthogonal then it can’t do much
- add new feature to active set
- compute new guess by solving OLS only with active features
Non-linear regression
- keep squared loss
- linear regression
- neural network with parameters
- true generative process
Two general approaches:
- transform features such that we can run linear regression (and use OLS)
- hand-crafted
- learned (first layer of a NN learns the parameters) - same as classification, except
- squared loss (instead of cross-entropy)
- last layers is linear (no activation)
- design algorithms to solve non-linear least squares
- if is not very high (), use Levenberg-Markquart
- regression trees and forests / Gaussian processes
- good results if is small (so NN cannot be trained)
I missed a few lectures here, the notes of the next chapter are taken from the recording of the last year’s “Fundamentals of Machine Learning class”.
Decision trees / forests
- idea: approximate a function using piecewise constant function
- in theory, we can get an arbitrarily good approximation
How to choose bins?
- classical: grid (great for , rather expensive for )
- better: define bins adaptively (bin size based on the number of datapoints)
- method: -means (defines bins by nearest-neighbour computation)
- method: recursive subdivision (what we’ll be doing)
For regression/classification, we can build a tree on top of it by doing the following:
Algorithm (generic tree building):
- create the root note with all data
- also put all of the data on the stack
- while the stack is not empty:
- take the top node
- check if the termination criterion is reached
- create a leaf node which stores the desired prediction
- otherwise
- find the best possible split and create 2 children
- put the data in the respective child
- put the children on the stack
For specific problems:
- termination criteria
- generic:
- stop at a defined depth (hyperparameter)
- stop at a defined number of instances in a node (hyperparameter)
- application-specific:
- classification: all instances in the node have the same label (then we don’t improve by splitting further) – we say they are pure
- generic:
- split criteria:
- axis-aligned, then decide left/right child by a 1-D threshold
- oblique splits (compute a new feature as a linear combination): sometimes more accurate but also more expensive
- we usually restrict possible thresholds to the midpoint between neighbouring instances – then for instances and features we have splits
- exhaustive search to find the best one (given a score function) or
- restrict the search to a random subset of features (eg. )
For classification trees:
- the prediction is the majority class in the leaf node
- the split criterion prefers splits that separate classes well… given a node :
- C4.5 algorithm: measure the entropy by iterating over all classes:
- CART algorithm: uses Gini impurity:
- in either case, the loss of a given split is (or if we use CART)
For classification trees:
- the prediction is the average value
- the split criterion minimizes the squared loss… given a node :
- the average is so we get
- the loss of a given split is the sum of the squared losses
Since trees tend to overfit, we can train several trees (i.e. forests). They have to be different for which there are a few tricks:
- in each node, consider a different random subset of features (we’re doing this)
- train each tree on a different random subset of the training set: bootstrapping:
- create a new by uniform sampling with replacement ( is initially empty, then add random instances)